-Search query
-Search result
Showing 1 - 50 of 91 items for (author: andreas & mp)

EMDB-41904: 
Klebsiella pneumoniae encapsulin-associated DyP peroxidase
Method: single particle / : Andreas MP, Jones JA, Giessen TW

EMDB-41905: 
Klebsiella pneumoniae DyP peroxidase-loaded encapsulin shell
Method: single particle / : Andreas MP, Jones JA, Giessen TW

EMDB-41906: 
Klebsiella pneumoniae SUMO-loaded encapsulin shell
Method: single particle / : Andreas MP, Jones JA, Giessen TW

EMDB-41078: 
Acinetobacter baumannii 118362 family 2A cargo-loaded encapsulin shell
Method: single particle / : Andreas MP, Benisch R, Giessen TW

EMDB-41322: 
Myxococcus xanthus EncA protein shell with compartmentalized SNAP-tag cargo protein
Method: single particle / : Andreas MP, Kwon S, Giessen TW

EMDB-29930: 
T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1
Method: single particle / : Schenk A, Deniston C, Noeske J

EMDB-28783: 
EsN-dhsU36mm2 full map (map 1)
Method: single particle / : Mueller AU, Chen J, Darst SA

EMDB-28784: 
EsN-dhsU36mm2 local map (map 2)
Method: single particle / : Mueller AU, Chen J, Darst SA

EMDB-28785: 
EsN-dhsU36mm2 composite map
Method: single particle / : Mueller AU, Chen J, Darst SA

EMDB-28786: 
EsN-dhsU36duplex full map (map 1)
Method: single particle / : Mueller AU, Chen J, Darst SA

EMDB-28790: 
EsN-dhsU36duplex local map (map 2)
Method: single particle / : Mueller AU, Chen J, Darst SA

EMDB-28791: 
EsN-dhsU36duplex composite map
Method: single particle / : Mueller AU, Chen J, Darst SA

EMDB-28792: 
EsN-dhsU36mm1 full map (map 1)
Method: single particle / : Mueller AU, Chen J, Darst SA

EMDB-28794: 
EsN-dhsU36mm1 local map (map 2)
Method: single particle / : Mueller AU, Chen J, Darst SA

EMDB-28797: 
EsN-dhsU36mm1 composite map
Method: single particle / : Mueller AU, Chen J, Darst SA

PDB-8f1i: 
SigN RNA polymerase early-melted intermediate bound to mismatch fragment dhsU36mm1 (-12T)
Method: single particle / : Mueller AU, Chen J, Darst SA

PDB-8f1j: 
SigN RNA polymerase early-melted intermediate bound to mismatch DNA fragment dhsU36mm2 (-12A)
Method: single particle / : Mueller AU, Chen J, Darst SA

PDB-8f1k: 
SigN RNA polymerase early-melted intermediate bound to full duplex DNA fragment dhsU36 (-12T)
Method: single particle / : Mueller AU, Chen J, Darst SA

EMDB-33329: 
High resolution cry-EM structure of the human 80S ribosome from SNORD127+/+ Kasumi-1 cells
Method: single particle / : Cheng J, Beckmann R

EMDB-33330: 
High resolution cry-EM structure of the human 80S ribosome from SNORD127+/- Kasumi-1 cells
Method: single particle / : Cheng J, Beckmann R

PDB-7xnx: 
High resolution cry-EM structure of the human 80S ribosome from SNORD127+/+ Kasumi-1 cells
Method: single particle / : Cheng J, Beckmann R

PDB-7xny: 
High resolution cry-EM structure of the human 80S ribosome from SNORD127+/- Kasumi-1 cells
Method: single particle / : Cheng J, Beckmann R

EMDB-27558: 
Acidipropionibacterium acidipropionici encapsulin in a closed state at pH 7.5
Method: single particle / : Jones JA, Andreas MP, Giessen TW

EMDB-27560: 
Acidipropionibacterium acidipropionici encapsulin in a closed state at pH 3.0
Method: single particle / : Jones JA, Andreas MP, Giessen TW

EMDB-27573: 
Acidipropionibacterium acidipropionici encapsulin in an open state at pH 7.5
Method: single particle / : Jones JA, Andreas MP, Giessen TW

EMDB-28669: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 01)
Method: electron tomography / : Liu J, Ren G
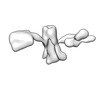
EMDB-28670: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 02)
Method: electron tomography / : Liu J, Ren G
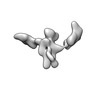
EMDB-28671: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 03)
Method: electron tomography / : Liu J, Ren G
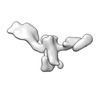
EMDB-28672: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 04)
Method: electron tomography / : Liu J, Ren G

EMDB-28673: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 05)
Method: electron tomography / : Liu J, Ren G
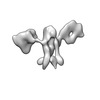
EMDB-28674: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 06)
Method: electron tomography / : Liu J, Ren G
EMDB-28675: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 07)
Method: electron tomography / : Liu J, Ren G
EMDB-28676: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 08)
Method: electron tomography / : Liu J, Ren G

EMDB-28677: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 09)
Method: electron tomography / : Liu J, Ren G

EMDB-28678: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 10)
Method: electron tomography / : Liu J, Ren G

EMDB-28679: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 11)
Method: electron tomography / : Liu J, Ren G

EMDB-28680: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 12)
Method: electron tomography / : Liu J, Ren G

EMDB-28681: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 13)
Method: electron tomography / : Liu J, Ren G

EMDB-28682: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 14)
Method: electron tomography / : Liu J, Ren G
EMDB-28683: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 15)
Method: electron tomography / : Liu J, Ren G
EMDB-28684: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 16)
Method: electron tomography / : Liu J, Ren G

EMDB-13926: 
Twist-corrected RNA origami 5-helix Tile A
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

PDB-7qdu: 
Twist-corrected RNA origami 5-helix Tile A
Method: single particle / : McRae EKS, Andersen ES

EMDB-13625: 
Mature conformer 2 of a 6-helix bundle of RNA with a clasp
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13626: 
Young conformer 2 of a 6-helix bundle of RNA with a clasp
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13627: 
6-Helix bundle of RNA
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13628: 
Young conformer of a 6-helix bundle of RNA with clasp
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13630: 
Mature conformer of a 6-helix bundle of RNA with clasp
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13633: 
RNA origami 5-helix tile
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13636: 
RNA origami 5-helix tile
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
